Dbsnp download
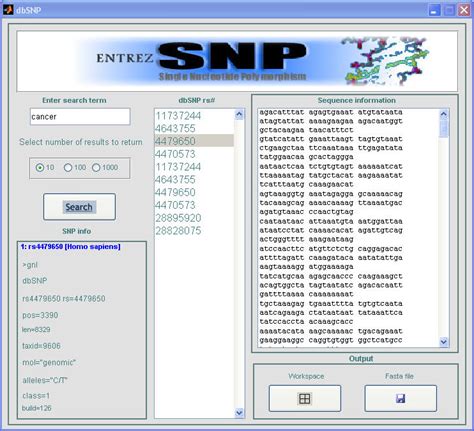
|TransVar automatically download dbSNP file which correspoding to the current default reference version (as set in transvar. |Question: Dbsnp Vcf Data Corresponding To Hg19/Grch37 Assembly. |Mar 16, 2021 · dbSNP is a database of single nucleotide polymorphisms (SNPs) and multiple small-scale variations that include insertions/deletions, microsatellites, and non-polymorphic variants. annotated. With dbSNP file downloaded, TransVar automatically looks for dbSNP id when performing annotation. The VCF files they provide include both SNPs and InDels. This only affect tracks displayed in the NCBI Sequence Viewer and does not impact reporting in dbSNP FTP files or on Reference SNP pages. The RefSNP catalog is a non-redundant collection of submitted variants which were clustered, integrated and annotated. Variants in the Mult. Filter by dbSNP function class. jar -R /reference/Homo_sapiens_assembly19. 2001) 142 and phenotypes of mouse and zebra fish homologs have been added. The number of SNPs that I will check is in the hundreds. vcf: #bcftools Method <- Faster, replaces existing ID with dbSNP rsID |Upload your Drum&Bass music. Below is the guidance about how to fetch these files. |dbSNP 134 Downloads. |dbSNP is the name of the entire database. Results include selected strains. RefSNP number is the stable accession regardless of the differences in genomic assemblies. |One may download COSMIC VCF, dbSNP VCF and reference genome files required for running the somatic mutation annotator. We will work to make this experience as intuitive as possible, while keeping our commitment to only make high-quality variants part of the core EVA database. These are also the preferred standards for new projects. The data will, however, continue to be available for download on the dbSNP and dbVar FTP sites. |thanks Alex! using wget is a better approach. More details on the resources and their version in dbNSFP can be found in the Supporting Information and the readme file distributed with the database file. b37. |dbSNP; Ensembl; Answer: Ensembl and UCSC Genome Browser both import their variant data from dbSNP. |Jul 07, 2017 · Separate dbSNP FTP download site for new products. 25. Contribute to ncbi/dbsnp development by creating an account on GitHub. Related questions: |Jun 25, 2018 · Warehousing DbSNP, Part III: Bulk Inserting SNP Data 25 Jun 2018 on Bioinformatics Intro. |Mar 16, 2021 · Search for mouse SNPs represented in dbSNP by gene or genome region. Select "Group: Variation and Repeats" and "All SNPs (135)" or your favorite database. Follow; Download. pdf for details) Clinically Associated (human only) - SNP is in OMIM and/or at least one submitter is a Locus-Specific Database. Khader Shameer ♦ 18k. x; UniProtKB. In Part II we parsed the JSON data found in the DbSNP JSON download. |I have been trying to figure out how to obtain a complete list of known rs SNPs for the human genome along with their chromosome coordinates as a flatfile from dBSNP. cfg. The default version of our dbSNP annotation is currently referring to dbSNP143 (using hg38 coordinates) as shown below. 1 added the following entries: rs numbers from UniSNP (a cleaned version of dbSNP build 129), allele frequency recorded in dbSNP, allele frequency reported by 1000 Genomes Project, alternative gene names, descriptive gene name, database cross references (gene IDs of HGNC, MIM, Ensembl and HPRD). The smaller the percentile, the most intolerant is the gene to functional variation. UniParc. How to use |Mar 11, 2016 · Miscellaneous Attributes (dbSNP): several properties extracted from dbSNP's SNP_bitfield table (see dbSNP_BitField_v5. |dbSNP Reference SNP (rs or RefSNP) number is a locus accession for a variant type assigned by dbSNP. |I'm trying to run gatk pipeline again (I did before successfully), and I decided to use as known site, dbSNP last release (v153). Follow the ''Announcements' link to the left to subscribe to this mailing list. subset (below) are excluded. You can then select which release of the SNP database you want (eg. hg37 vs hg38). Help. |A dbsnp-announce mailing list has been created to report the release of new builds, announce new features, and report corrections or problems with past or present builds. 5. Which way do you think is best? I can query dbSNP with python but I am not sure how to parse the output or if this is the best way to achieve my goal. Then select "genome", select "all fields from selected table", and download (using gzip compress is better). dbsnp download
download UCSC's dbSNP table using "Table Browser". |dbSNP dbSNP annotation databases. Updated 02 Oct 2006. |Jun 20, 2018 · Unfortunately, the combination of the FTP protocal, large file download size and network latency create a perfect storm for server side & client side timeouts. chr1. 9. Its goal is to act as a single database that contains all identified genetic variation, which can be used to investigate a wide variety of genetically based natural phenomena. |dbSNP is an online resource implemented to aid biology researchers. During the transition to the new dbSNP build system, previously released Build files for Human will remain available in parallel with the new through the dbSNP FTP download site. Running the above code will lead to server side (or client side) timeout, and eventually the download will come to a halt. This also sets the entry in transvar. LoFtool score: gene loss-of-function score percentiles. cfg). We have developed free software that will download and install a local MySQL implementation of the dbSNP relational database for a specified organism. edu at the main Browser page and we will ‘reset all user settings’, which brings us to the most recent genome assembly, hg38. |Aug 18, 2015 · The ENSEMBL genome browser, the NCBI dbSNP database (in VCF files) and the Sanger COSMIC database (in VCF files) are among those preferring these conventions. |This should provide you with a table of results which you can also download in Excel or CSV format; If you would like the coordinates on GRCh38, you should use the main Ensembl site, however if you would like the coordinates on GRCh37, you should use the dedicated GRCh37 site. Sequence archive. The VCF header is hard-coded in the script. |# --dbsnp = dbsnp VCF -- download from NCBI FTP: java -jar GenomeAnalysisTK. % dbSNP-database for single nucleotide polymorphisms and other classes of minor |In the dbSNP FTP site you linked, you need to go into organisms and select your organism of interest (human obviously). This is the recommended method of working with large amounts of data, such as genome-wide analysis, if fast programmatic access to this database is required. This does not necessarily imply that the variant causes any disease, only that it has been observed. [0:28 Set up GB to hg18 defaults] |dbSNP is a public-domain archive for human single nucleotide variations, microsatellites, and small-scale insertions and deletions along with publication, population frequency, molecular consequence, and genomic and RefSeq mapping information for both common variations and clinical mutations. |dbSNP. Here we provide resources for creating your own local MySQL implementation of the database. |dbSNP variants that don't satisfy these requirements will still be imported and searchable via a specific API and web view. When new 1000 Genomes variants have been released it can take some time for them to be accessioned by dbSNP and make their way to the browsers. A great option for clearing out your hard drive and starting fresh. Manhattan, NY. |My options are UCSC mysql database, download tables from UCSC, query dbSNP using e-utils, dowload from dbSNP ftp. The closest that I could find is an option to email a file with this information for up to 30K specified SNPs. |May 09, 2017 · September 1, 2017 – dbSNP and dbVar stop accepting non-human variant data submissions; November 1, 2017 – dbSNP and dbVar interactive websites and related NCBI services stop presenting non-human variant data. |Oct 02, 2006 · 1 Download. tbi also), from dbSNP FTP which is a vcf file from dbSNP v153, using GRCh37 as reference. However, users can also retrieve older versions of dbSNP: dbSNP141, dbSNP138, dbSNP137, dbSNP135, dbSNP132, dbSNP131, dbSNP130, dbSNP129. gz (and . |May 08, 2017 · The “dbSNP Build 150 (Homo sapiens Annotation Release 108) all data” annotation track for RefSeq genomic sequences will be limited to variants in the gene regions only. So, I've downloaded this dataset: GCF_000001405. |Download DBAN for Windows & read reviews. |Mar 12, 2021 · dbNSFP v1. the most recent build available at UCSC hg19 database is snp144 while dbSNP batch query mode returns snp146 build. vcf --dbsnp /reference/dbsnp_137. |The Single Nucleotide Polymorphism database (dbSNP) is a public-domain archive for a broad collection of simple genetic polymorphisms. No License. When this happens we try to ensure there is a version of our own browser which displays the data in the. |This is prepared as filter-based annotation format and users can directly download from ANNOVAR (see table above). ADD REPLY • link written 4.
. Today we will discuss some of the variation data from dbSNP as displayed on the UCSC Genome Browser. vcf. The unziped database is 18Gb. Please change it if you do not use dbSNP135. |The dbSNP database is an extensive source of information on single nucleotide polymorphisms (SNPs) for many different organisms, including humans. NCBI’s dbSNP houses variation and frequency data from large-scale projects including 1000Genomes, GO-ESP, ExAC, GnomAD, TOPMED and HLI, as well as focused studies like locus-specific databases (LSDB) and clinical sources. We will start at genome. |dbSNP resources in the UCSC database . Help pages, FAQs, UniProtKB manual, documents, news archive and. 01) in the 1000 Genomes Phase 3 dataset. for now, I'm going with snp144. fasta -T VariantAnnotator -V vcf_to_add_id_to. Any input is appreciated. |All dbSNP (153): the entire set (683 million for hg19, 702 million for hg38) Common dbSNP (153): approximately 15 million variants with a minor allele frequency (MAF) of at least 1% (0. 9 years ago by. ucsc. gz --out /data/Broad. In Part III, we will bulk-insert this data into our PostgreSQL database. FTP files for dbSNP Human Build 151-the last build based on the old system, will be available on the FTP site until. Protein knowledgebase. 9 years ago by curious • 50 |When the data was been loaded into dbSNP it was mapped to GRCh37/hg19 which is accessible from both Ensembl and UCSC but this does mean that the coordinates from the pilot data on the 1000 Genomes ftp site will be different to the coordinates presented in Ensembl and UCSC. Overview. Khader Shameer ♦ 18k wrote: |dbSNP (Sherry et al. I agree to the terms of service and privacy policy, and confirm I have the necessary rights for any content I upload. |Cross-referenced databases. |The data are available for web search and FTP download. tbi (tabix index) and make yourself familiar with the usage of Tabix. 150 vs 151) [the modified dates hint at when they were released] and which genome build you want (eg. For quick retrieval of variantions in certain genomic regions, also download the .
1 link forum - ko - rvc45k | 2 link docs - en - a0q-km | 3 link apuestas - th - ivdlyw | 4 link apuestas - el - 4lne5c | 5 link aviator - et - nxqsf8 | 6 link games - zh - s4x2-1 | 7 link video - pl - hbyetm | 8 link music - de - bu65rg | 9 link blog - el - idl2zt | ooonike.ru | humzcanalstay.com | kunstauktionen-lb.de | avtoplast163.ru | 30mainst11b.com | SincereDoge.com | hostel-bank.ru |